Fixation Before Dissociation Using a Deep Eutectic Solvent Preserves In Vivo States and Phospho-Signalling in Single-Cell Sequencing Presented
Researchers at the University of Alabama at Birmingham have demonstrated a novel way of fixing cells and preserving their biomolecules prior to single cell dissociation without the need for harsh enzymatic/mechanical approaches.
Single-cell RNA sequencing has the opportunity to deconstruct cellular networks but was previously limited by the loss of biological information caused by traditional dissociation methods. vivoPHIX™, a Deep Eutectic Solvent (DES) is a non-cross linking reagent that rapidly fixes bacterial, plant and animal cells, allowing the long-term stabilisation of biomolecules and facilitating single cell dissociation, as demonstrated in this recent bioRxiv publication.
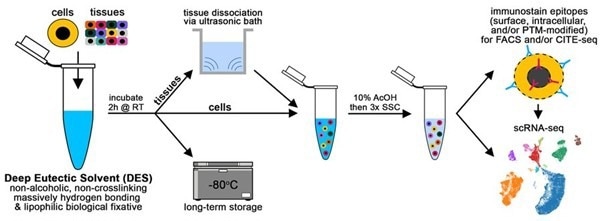
Fig 1. Workflow demonstrating cell/tissue fixation using a DES (vivoPHIX™) for CITE-Seq and scRNA-Seq.
The research in this paper demonstrates:
- Long term stabilisation of biobanked tissues, resulting in high-quality scRNA-seq data without any activation artifacts using vivoPHIX™.
- Complex cellular morphologies are retained with ultrasonic dissociation, such as neurons of the human retina
- Post-translational modifications are preserved, providing sufficient membrane permeabilization for probing intracellular epitopes.
We anticipate that DES-based fixatives will allow the reconstruction of in vivo cellular networks & uncover cooperativity b/w intracellular pathways. We believe that DES could really change the field of single-cell analysis’.
Seth Fortmann, Main Author
Andrew Goldsborough, CEO at RNAssist Ltd who developed vivoPHIX™ and supported the project agrees. ‘This is a herculean effort by Seth and the team at UAB, with so much data produced it could warrant three separate publications. It’s a true testament to Seth’s hard work over the past 3 years and highlights the enormous potential vivoPHIX™ has in the single-cell world.’
For further information or to order vivoPHIX™ please contact [email protected]
Source: Read Full Article




